The immune system is our body’s frontline defense, protecting us from bacteria, viruses, and other microbes. It relies on multiple layers of protection — from specialized “adaptive” responses, such as antibodies and T cells that recognize specific threats, to rapid “innate” mechanisms that respond to common features of microbes, as well as physical barriers in the skin, lungs and gut that help prevent infection.
Researchers in the Lacy-Hulbert Lab study how these components of the immune system work together to defend against harmful invaders while avoiding damage to healthy tissues or harmless microbes. Understanding this balance can reveal new strategies to prevent and treat immune system diseases.
Support for the lab comes from the National Institutes of Health, the Crohn’s and Colitis Foundation, Wellcome, the Lupus Research Alliance, the Seattle Foundation, and other partners.

Adam Lacy-Hulbert, PhD
Lab Members

Kayla Fasano

Caitlyn Kwong

Jane Madden

Kelsey Mauk, PhD

Caroline Stefani, PhD
Lauren Vandepas, PhD

Anna Yoshida

Cheng Zhao, PhD
Research Projects
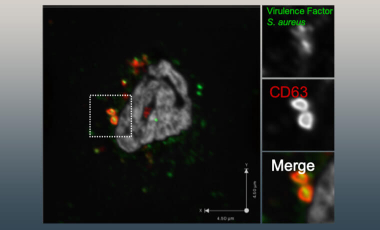
Distinguishing Pathogens From Self
Researchers in the Lacy-Hulbert Lab are exploring the molecular and cellular pathways that determine when innate immune cells promote tolerance and when they trigger inflammation.
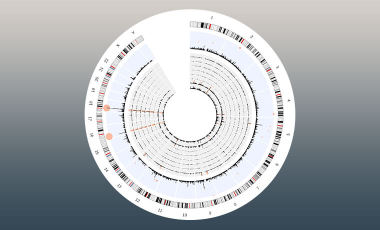
Forward Genetics To Identify New Mechanisms in Immunity and Host Defense
In the Lacy-Hulbert Lab, researchers focus on identifying the mechanisms that control innate immunity and host defense, using innovative genetic approaches that let them explore immunity on an unprecedented scale.

Regulation of Immune Responses
The Lacy-Hulbert Lab seeks to understand how the immune system maintains balance — mounting strong defenses against infection while avoiding harmful attacks on the body itself.
SARS-CoV2 And COVID-19
During the COVID-19 pandemic, the Lacy-Hulbert Lab applied its expertise to better understand SARS-CoV-2, the virus that causes COVID-19, and explore ways to prevent and treat infection.