Autoimmunity is controlled in healthy individuals by cells that actively control immune responses to self so that the body is tolerant of it’s own tissues. The Long Lab focuses on understanding mechanisms of tolerance, why they are lost in autoimmunity, and how tolerance can be augmented with therapy.
Our approach combines in-depth characterization of T cells that confer tolerance, discovery of drivers of tolerance, and testing of how therapies modulate tolerance in trials. We use cutting-edge single cell assays to assess rare antigen-specific responses in clinical samples, with a focus on type 1 diabetes and related autoimmune diseases. These approaches are complemented by work in the Human Immunophenotyping Core.
Our goal is to better treat and prevent autoimmunity by augmenting T cell tolerance.

S. Alice Long, PhD
Lab Members

Christine Bender, PhD

Justine Calise, PhD

Sarah Kobernat, PhD

Alice Wiedeman, PhD
Past Lab Members
Long Lab
- Valerie Wall – Notch Therapeutics
- Rebecca LaFond – Mason University
- Duangchan Suwannasaen – Flow Contract Site
- Katharine Schwedhelm – Fred Hutch
HIP Core
- Lori Blanchfeild – Alpine Immune Sciences
- Bryce Fuchs – University of Utah
- Jerill Thorpe – Bristol Myers Squibb
- Megan Maers – University of Washington
- Nathan Alidina – AGC Biologics
- Ian Frank – Sony Biotechnology
Research Projects
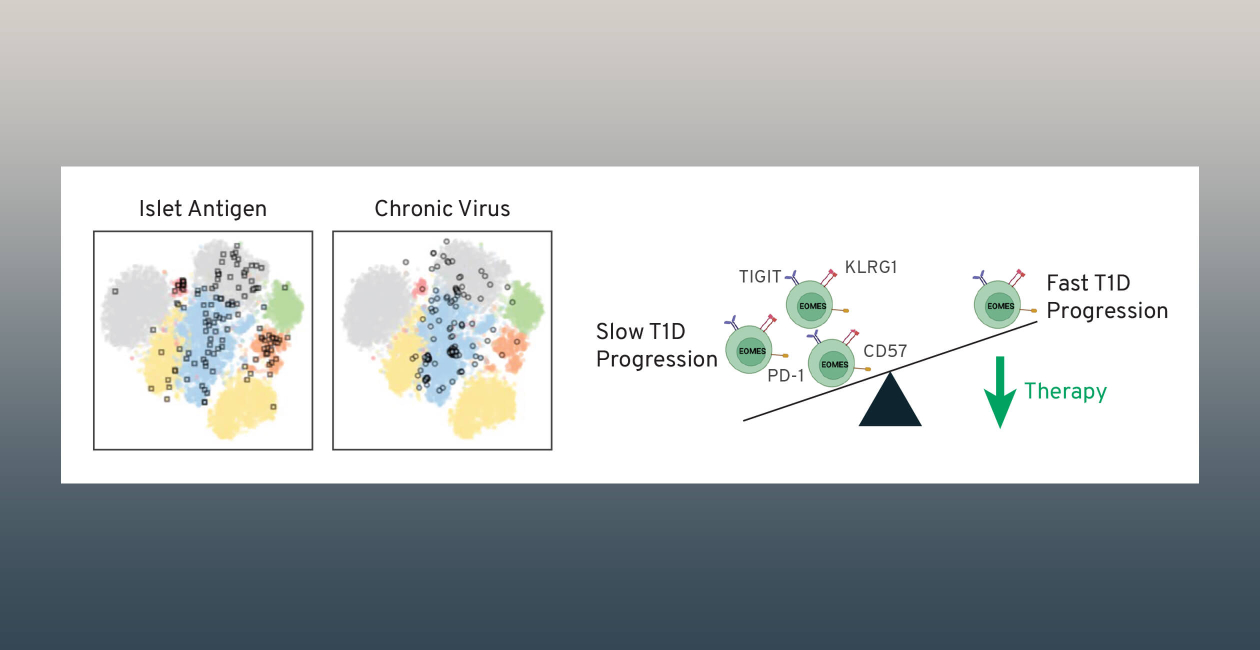
Drivers of CD8 T cells exhaustion in autoimmunity
Impact of IL-2/IL-2R signaling in T1D
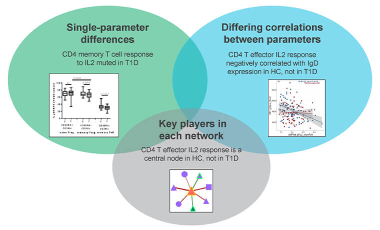
Mechanisms of tolerance with autoimmune therapies
Featured Publications
-
Feb 2021
Exhausted-like CD8+ T cell phenotypes linked to C-peptide preservation in alefacept-treated T1D subjects.
JCI InsightDiggins KE, Serti E, Muir V, Rosasco M, Lu T, Balmas E, Nepom GT, Long SA, Linsley PS -
Jan 2020
Autoreactive CD8+ T cell exhaustion distinguishes subjects with slow type 1 diabetes progression.
J Clin InvestWiedeman AE, Muir VS, Rosasco MG, DeBerg HA, Presnell S, Haas B, Dufort MJ, Speake C, Greenbaum CJ, Serti E, Nepom GT, Blahnik G, Kus AM, James EA, Linsley PS, Long SA -
Aug 2019
An Anti-CD3 Antibody, Teplizumab, in Relatives at Risk for Type 1 Diabetes.
N Engl J MedHerold KC, Bundy BN, Long SA, Bluestone JA, DiMeglio LA, Dufort MJ, Gitelman SE, Gottlieb PA, Krischer JP, Linsley PS, Marks JB, Moore W, Moran A, Rodriguez H, Russell WE, Schatz D, Skyler JS, Tsalikian E, Wherrett DK, Ziegler AG, Greenbaum CJ, Type 1 Diabetes TrialNet Study Group. -
Jun 2019
Dynamic Immune Phenotypes of B and T Helper Cells Mark Distinct Stages of T1D Progression.
DiabetesHabib T, Long SA, Samuels PL, Brahmandam A, Tatum M, Funk A, Hocking AM, Cerosaletti K, Mason MT, Whalen E, Rawlings DJ, Greenbaum CJ, Buckner JH, Type 1 Diabetes TrialNet Study Group. -
Nov 2016
Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes.
Sci ImmunolLong SA, Thorpe J, DeBerg HA, Gersuk V, Eddy J, Harris KM, Ehlers M, Herold KC, Nepom GT, Linsley PS