Dr. Cerosaletti’s research is focused on the role of the adaptive immune system in the development and progression of immune mediated diseases and the response to treatment.
Current research is examining the role of T cells in the development and progression of autoimmune type 1 diabetes (T1D), and the response to immunotherapy in T1D. The lab is identifying transcriptional and T cell receptor signatures of islet autoreactive CD4 T effector cells and regulatory T cells that are linked to disease course.
The Cerosaletti lab is also examining the expansion of stem-like CD4 memory T cells in T1D which may serve as a reservoir of autoreactive T cells in T1D.
Additionally, Dr. Cerosaletti has a longstanding research interest in the molecular genetics of immune disorders to establish functional links between genetic variants and alterations in the immune response leading to loss of tolerance, viral response and response to therapy. Current work is focused on the impact of genetic variants in the response to peanut allergy immunotherapy.

Karen Cerosaletti, PhD
Research Projects
Autoreactive T cells in type 1 diabetes
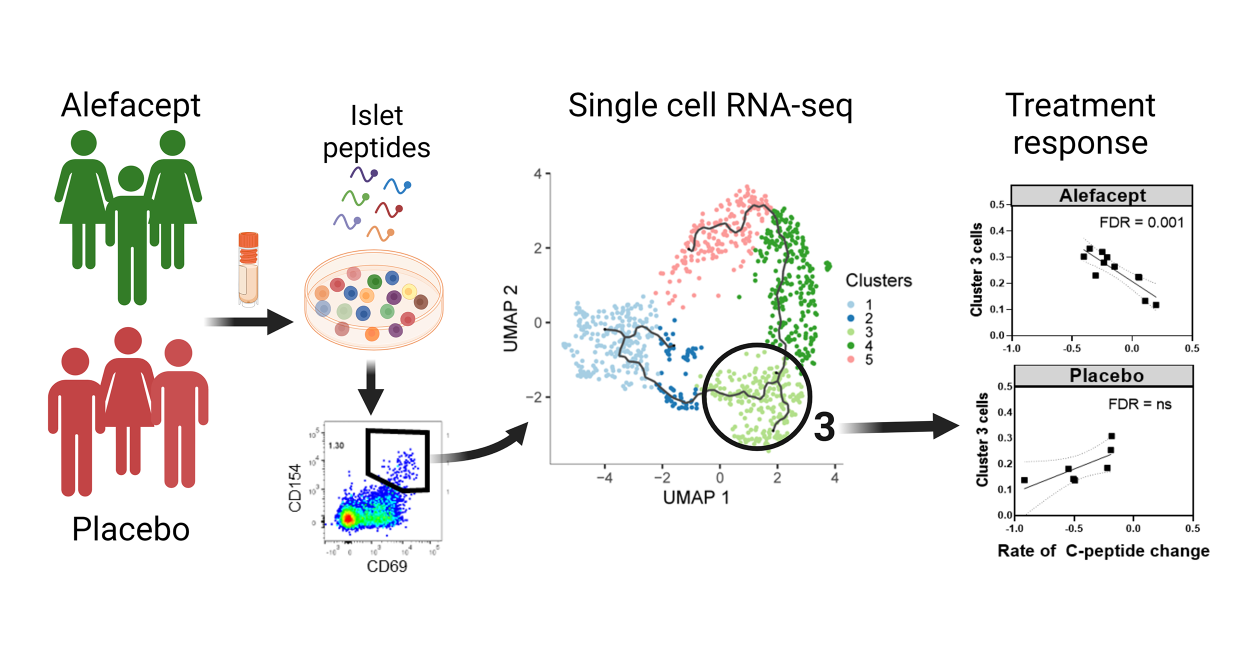
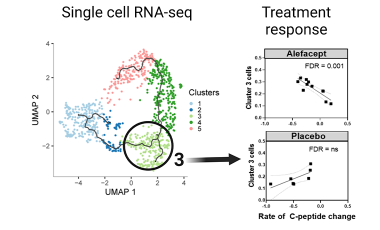
The lab is investigating autoreactive effector and regulatory T cells in T1D using single cell RNA sequencing to identify features of these cells that are linked to disease course or response to therapy.
Genetics of response to allergy immunotherapy
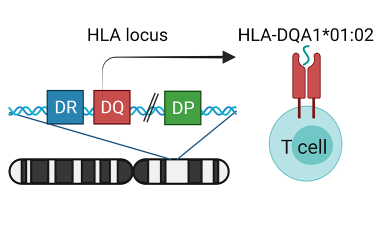
The Cerosaletti Lab is studying the role of genetic variation in the HLA class II genes in the response to peanut oral immunotherapy by analysis of antigen presentation and recognition by peanut specific T cells.
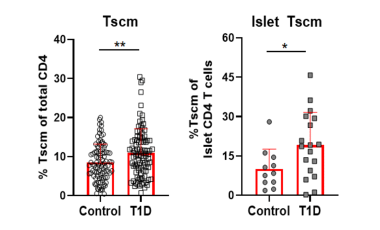
Stem-like memory CD4 T cells in type 1 diabetes
The Cerosaletti lab is studying stem-like memory CD4 T cells (Tscm) in T1D to understand the mechanism leading to expansion of these cells in T1D and determine if Tscm levels influence disease progression and response to therapy.