Genetic variation in the human genome controls a compendium of cellular and physiological components that make up human biology. The Ray Lab is interested in better understanding how genetic variation tunes our immune system and how this can put individuals at risk for autoimmune diseases, which is a vital step in developing more effective therapeutics with fewer side effects.
Because most genetic risk for autoimmune diseases occurs in non-coding regions of the genome, the Ray lab studies how non-coding genetic variants modulate cis-regulatory regions and alter immune cell activities that lead to autoimmune disease susceptibility. We prioritize likely disease-causal variants and cis-regulatory regions, and, in human and mouse systems, identify variant target genes and pathways and define their functional effects on immune cells.
These studies will inform efforts for personalized therapies and disease prevention.


John Ray, PhD
Lab Members

Alex Ho, PhD

Hannah Kalinoski, PhD

Lucy Li

Meghan McQuade

LeAnn Nguyen, PhD

Amelia Querbach

Stephanie Ryder

Oscar Sias-Garcia

Marlana Winschel
Research Projects
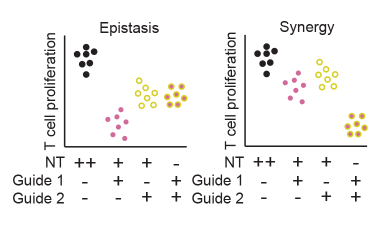
Defining variants that act collectively to promote disease
Identifying synergistic properties of variants and enhancers on T cell function
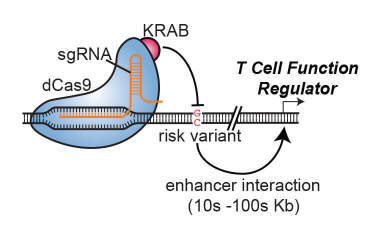
Mapping cis-regulatory regions that control immune cell function genome-wide
Identifying enhancers that facilitate T and B cell function throughout the genome using CRISPR screening approaches.
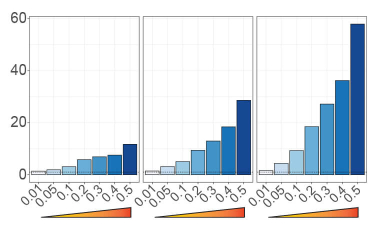
Prioritizing autoimmune disease-causal variants in immune cells
Utilizing massively parallel reporter assays and statistical fine-mapping to enrich for functional and disease-causal genetic variation