Genome-wide association studies (GWAS) have associated thousands of genetic loci with commonly acquired autoimmune diseases. However, the genetic variants that contribute to autoimmunity in each locus are mostly unknown. This is because disease contributing variants are often in tight linkage disequilibrium with many non-contributing variants, and 90% of disease-associated variants are in non-coding regions, making their actions on gene expression and cellular processes difficult to measure. One clue for disease variant mechanisms of action is their enrichment in gene regulatory regions called enhancers.
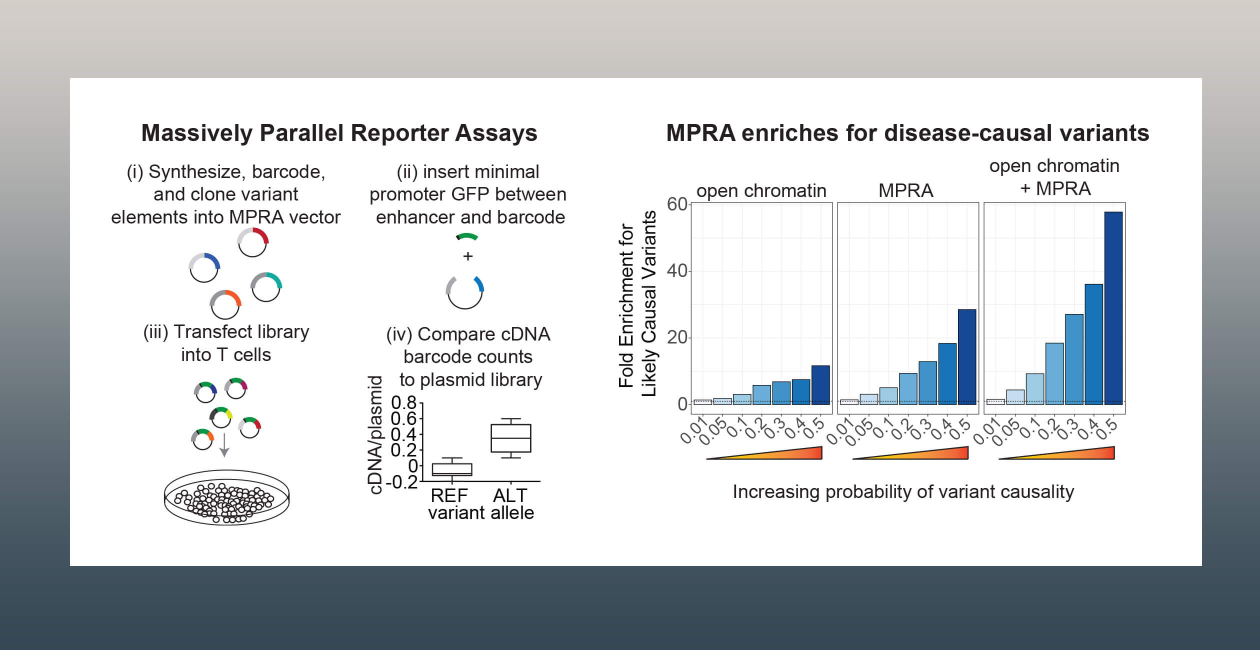
Our lab utilizes massively parallel reporter assays (MPRA) to test genetic variants for their effects on enhancer activity in a plasmid-based reporter system. Using this system, we can test tens of thousands of genetic variants for their effects on enhancer activity.
Recently, we found that variants that have allele-specific differences in MPRA conducted in a T cell line enrich highly, up to 60-fold, for disease-causal genetic variants for 5 autoimmune diseases. Thus, we have found MPRA to be a vital tool for identifying complex trait causal variants that alter enhancer activity. The lab’s current aim is to scale this to identify causal variants that regulate all immune cell traits.
Additional Research Projects
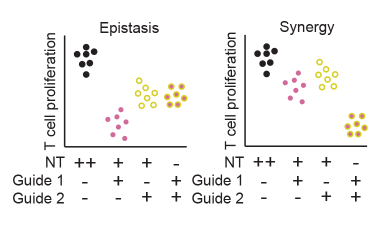
Defining variants that act collectively to promote disease
Identifying synergistic properties of variants and enhancers on T cell function
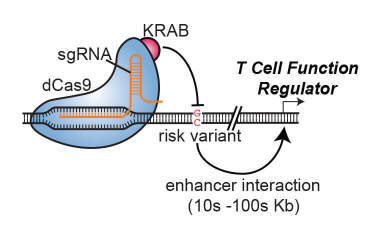
Mapping cis-regulatory regions that control immune cell function genome-wide
Identifying enhancers that facilitate T and B cell function throughout the genome using CRISPR screening approaches.